-
Duan Chen
Our recent work is motivated by two types of biological problems. One is inferring 3D structures of chromatins based on chromosome conformation capture (3C), such as Hi-C, which is a high-throughput sequencing technique that produces millions of contact data between genomic loci pairs. The other problem is computational deconvolution of gene expression data from heterogeneous brain samples, for extracting cell type-specific information for patients with Alzheimer's Disease (AD). Both problems involve large volumes of data, thus fast algorithms are indispensable in either direct optimization or machine learning methods. A central approach is the low-rank approximation of matrices. Conventional matrix decomposition methods such as SVD, QR, etc, are expensive, so not suitable for repeated implementation in these biological problems. Instead, we develop fast stochastic matrix compressions based on randomized numerical linear algebra (RNLA) theories. In this talk, we will emphasize on a recently developed stochastic kernel matrix compression algorithm. In this algorithm, samples are taken at no (or low) cost and the original kernel matrix is reconstructed efficiently with desired accuracy. Storage and compressing processes are only at O(N) or O(NlogN) complexity. These stochastic matrix compressing can be used to the above-mentioned biological problems to greatly improve algorithm efficiency, they can also be applied to other kernel based machine learning algorithms for scientific computing problems with non-local interactions (such as fractional differential equations), since no analytic formulation of the kernel function is required in our algorithms.
-
Duan Chen
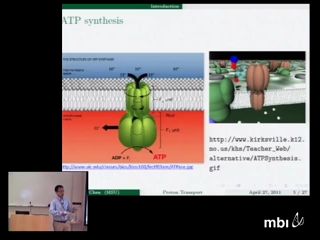
Proton transport across membranes is one of the most important and interesting phenomena in living cells. The present work proposes a multiscale/multiphysical model for the understanding of atomic level mechanism of proton transport in transmembrane proteins. We describe proton dynamics quantum mechanically via a density functional approach while implicitly model numerous solvent molecules as a dielectric continuum to reduce the number of degrees of freedom. The impact of protein molecular structure and its charge polarization on the proton transport is considered explicitly in atomic level. The molecular surface of the channel protein is utilized to split the discrete protein domain and the continuum solvent domain, and facilitate the multiscale discrete/continuum/quantum descriptions. We formulate a total free energy functional to put proton kinetic and potential energies as well as electrostatic energy of all ions on an equal footing. Generalized Poisson-Boltzmann equation and Kohn-Sham equation are obtained from the variational framework. A number of mathematical algorithms, including the Dirichlet to Neumann mapping, matched interface and boundary method, Gummel iteration, and Krylov space techniques are utilized to implement the proposed model in a computationally efficient manner. The Gramicidin A (GA) channel is used to demonstrate the performance of the proposed proton channel model and validate the efficiency of the proposed mathematical algorithms. The electrostatic characteristics and proton conductance of the GA channel are analyzed with a wide range of model parameters. A comparison with experimental data verifies the present model predictions and validates the proposed model.
-
Duan Chen
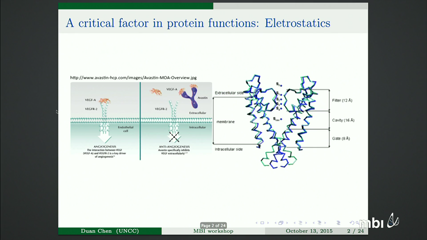
Description of inhomogeneous dielectric properties of a solvent in the vicinity of ions has been attracting research interests in mathematical modeling for many years. From many experimental results, it has been concluded that the dielectric response of a solvent linearly depends on the ionic strength within a certain range. Based on this assumption, a new implicit solvent model is proposed in the form of total free energy functional and a quasi-linear Poisson-Boltzmann equation (QPBE) is derived. Classical Newton’s iteration can be used to solve the QPBE numerically but the corresponding Jacobian matrix is complicated due to the quasi-linear term. In the current work, a systematic formulation of the Jacobian matrix is derived. As an alternative option, an algorithm mixing the Newton’s iteration and the fixed point method is proposed to avoid the complicated Jacobian matrix, and it is a more general algorithm for equation with discontinuous coefficients. Computational efficiency and accuracy for these two methods are investigated based on a set of equation parameters. At last, the QPBE with singular charge source and piece-wisely defined dielectric functions has been applied to analyze electrostatics of macro biomolecules in a complicated solvent. A set of computational algorithms such as interface method, singular charge removal technique and the Newton- fixed-point iteration are employed to solve the QPBE. Biological applications of the proposed model and algorithms are provided, including calculation of electrostatic solvation free energy of proteins, investigation of physical properties of channel pore of an ion channel, and electrostatics analysis for the segment of a DNA strand.
 Duan ChenOur recent work is motivated by two types of biological problems. One is inferring 3D structures of chromatins based on chromosome conformation capture (3C), such as Hi-C, which is a high-throughput sequencing technique that produces millions of contact data between genomic loci pairs. The other problem is computational deconvolution of gene expression data from heterogeneous brain samples, for extracting cell type-specific information for patients with Alzheimer's Disease (AD). Both problems involve large volumes of data, thus fast algorithms are indispensable in either direct optimization or machine learning methods. A central approach is the low-rank approximation of matrices. Conventional matrix decomposition methods such as SVD, QR, etc, are expensive, so not suitable for repeated implementation in these biological problems. Instead, we develop fast stochastic matrix compressions based on randomized numerical linear algebra (RNLA) theories. In this talk, we will emphasize on a recently developed stochastic kernel matrix compression algorithm. In this algorithm, samples are taken at no (or low) cost and the original kernel matrix is reconstructed efficiently with desired accuracy. Storage and compressing processes are only at O(N) or O(NlogN) complexity. These stochastic matrix compressing can be used to the above-mentioned biological problems to greatly improve algorithm efficiency, they can also be applied to other kernel based machine learning algorithms for scientific computing problems with non-local interactions (such as fractional differential equations), since no analytic formulation of the kernel function is required in our algorithms.
Duan ChenOur recent work is motivated by two types of biological problems. One is inferring 3D structures of chromatins based on chromosome conformation capture (3C), such as Hi-C, which is a high-throughput sequencing technique that produces millions of contact data between genomic loci pairs. The other problem is computational deconvolution of gene expression data from heterogeneous brain samples, for extracting cell type-specific information for patients with Alzheimer's Disease (AD). Both problems involve large volumes of data, thus fast algorithms are indispensable in either direct optimization or machine learning methods. A central approach is the low-rank approximation of matrices. Conventional matrix decomposition methods such as SVD, QR, etc, are expensive, so not suitable for repeated implementation in these biological problems. Instead, we develop fast stochastic matrix compressions based on randomized numerical linear algebra (RNLA) theories. In this talk, we will emphasize on a recently developed stochastic kernel matrix compression algorithm. In this algorithm, samples are taken at no (or low) cost and the original kernel matrix is reconstructed efficiently with desired accuracy. Storage and compressing processes are only at O(N) or O(NlogN) complexity. These stochastic matrix compressing can be used to the above-mentioned biological problems to greatly improve algorithm efficiency, they can also be applied to other kernel based machine learning algorithms for scientific computing problems with non-local interactions (such as fractional differential equations), since no analytic formulation of the kernel function is required in our algorithms. Duan ChenProton transport across membranes is one of the most important and interesting phenomena in living cells. The present work proposes a multiscale/multiphysical model for the understanding of atomic level mechanism of proton transport in transmembrane proteins. We describe proton dynamics quantum mechanically via a density functional approach while implicitly model numerous solvent molecules as a dielectric continuum to reduce the number of degrees of freedom. The impact of protein molecular structure and its charge polarization on the proton transport is considered explicitly in atomic level. The molecular surface of the channel protein is utilized to split the discrete protein domain and the continuum solvent domain, and facilitate the multiscale discrete/continuum/quantum descriptions. We formulate a total free energy functional to put proton kinetic and potential energies as well as electrostatic energy of all ions on an equal footing. Generalized Poisson-Boltzmann equation and Kohn-Sham equation are obtained from the variational framework. A number of mathematical algorithms, including the Dirichlet to Neumann mapping, matched interface and boundary method, Gummel iteration, and Krylov space techniques are utilized to implement the proposed model in a computationally efficient manner. The Gramicidin A (GA) channel is used to demonstrate the performance of the proposed proton channel model and validate the efficiency of the proposed mathematical algorithms. The electrostatic characteristics and proton conductance of the GA channel are analyzed with a wide range of model parameters. A comparison with experimental data verifies the present model predictions and validates the proposed model.
Duan ChenProton transport across membranes is one of the most important and interesting phenomena in living cells. The present work proposes a multiscale/multiphysical model for the understanding of atomic level mechanism of proton transport in transmembrane proteins. We describe proton dynamics quantum mechanically via a density functional approach while implicitly model numerous solvent molecules as a dielectric continuum to reduce the number of degrees of freedom. The impact of protein molecular structure and its charge polarization on the proton transport is considered explicitly in atomic level. The molecular surface of the channel protein is utilized to split the discrete protein domain and the continuum solvent domain, and facilitate the multiscale discrete/continuum/quantum descriptions. We formulate a total free energy functional to put proton kinetic and potential energies as well as electrostatic energy of all ions on an equal footing. Generalized Poisson-Boltzmann equation and Kohn-Sham equation are obtained from the variational framework. A number of mathematical algorithms, including the Dirichlet to Neumann mapping, matched interface and boundary method, Gummel iteration, and Krylov space techniques are utilized to implement the proposed model in a computationally efficient manner. The Gramicidin A (GA) channel is used to demonstrate the performance of the proposed proton channel model and validate the efficiency of the proposed mathematical algorithms. The electrostatic characteristics and proton conductance of the GA channel are analyzed with a wide range of model parameters. A comparison with experimental data verifies the present model predictions and validates the proposed model. Duan ChenDescription of inhomogeneous dielectric properties of a solvent in the vicinity of ions has been attracting research interests in mathematical modeling for many years. From many experimental results, it has been concluded that the dielectric response of a solvent linearly depends on the ionic strength within a certain range. Based on this assumption, a new implicit solvent model is proposed in the form of total free energy functional and a quasi-linear Poisson-Boltzmann equation (QPBE) is derived. Classical Newton’s iteration can be used to solve the QPBE numerically but the corresponding Jacobian matrix is complicated due to the quasi-linear term. In the current work, a systematic formulation of the Jacobian matrix is derived. As an alternative option, an algorithm mixing the Newton’s iteration and the fixed point method is proposed to avoid the complicated Jacobian matrix, and it is a more general algorithm for equation with discontinuous coefficients. Computational efficiency and accuracy for these two methods are investigated based on a set of equation parameters. At last, the QPBE with singular charge source and piece-wisely defined dielectric functions has been applied to analyze electrostatics of macro biomolecules in a complicated solvent. A set of computational algorithms such as interface method, singular charge removal technique and the Newton- fixed-point iteration are employed to solve the QPBE. Biological applications of the proposed model and algorithms are provided, including calculation of electrostatic solvation free energy of proteins, investigation of physical properties of channel pore of an ion channel, and electrostatics analysis for the segment of a DNA strand.
Duan ChenDescription of inhomogeneous dielectric properties of a solvent in the vicinity of ions has been attracting research interests in mathematical modeling for many years. From many experimental results, it has been concluded that the dielectric response of a solvent linearly depends on the ionic strength within a certain range. Based on this assumption, a new implicit solvent model is proposed in the form of total free energy functional and a quasi-linear Poisson-Boltzmann equation (QPBE) is derived. Classical Newton’s iteration can be used to solve the QPBE numerically but the corresponding Jacobian matrix is complicated due to the quasi-linear term. In the current work, a systematic formulation of the Jacobian matrix is derived. As an alternative option, an algorithm mixing the Newton’s iteration and the fixed point method is proposed to avoid the complicated Jacobian matrix, and it is a more general algorithm for equation with discontinuous coefficients. Computational efficiency and accuracy for these two methods are investigated based on a set of equation parameters. At last, the QPBE with singular charge source and piece-wisely defined dielectric functions has been applied to analyze electrostatics of macro biomolecules in a complicated solvent. A set of computational algorithms such as interface method, singular charge removal technique and the Newton- fixed-point iteration are employed to solve the QPBE. Biological applications of the proposed model and algorithms are provided, including calculation of electrostatic solvation free energy of proteins, investigation of physical properties of channel pore of an ion channel, and electrostatics analysis for the segment of a DNA strand.